Translational Research Departments
The clinicians and scientists at the Saint John’s Cancer Institute work collaboratively to advance patient care. Our scientists investigate the mechanisms underlying a disease target of interest, and work directly with clinicians to translate lab-based discoveries into personalized treatments. This bench-to-beside approach fosters a truly beneficial experience for our patients, as they lead to novel findings and new directions for therapeutic interventions.



Translational Molecular Medicine
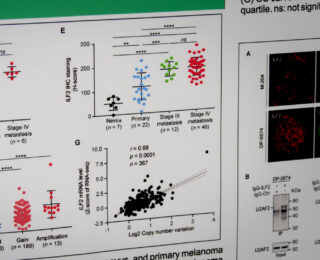
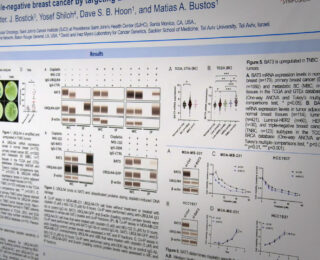
The Department of Translational Molecular Medicine is committed to the development and translational application of transcriptomic, genomic, and epigenomic biomarkers as diagnostic, prognostic and predictive tools in patients with solid tumors, particularly melanoma, breast cancer, prostate cancer, and gastrointestinal tract cancers. With our academic and industries collaborators, we develop quantitative translational oncology tools to improve management of solid tumor cancer patients. We also co-develop treatment protocols and new molecular oncology approaches, working to increase survival rates and promote a cure for patients with solid tumor cancers.
Learn More
Translational Neurosciences
The department of Translational Neurosciences is accelerating the development of novel therapies for brain cancers and neurological disorders through drug innovations and precision medicine. Clinical trials for neuroscience including; brain cancer, brain metastasis, solid tumors, neurology, as well as quality and outcomes research. The Translational Neurosciences also collaborates with various organizations including; The Institute for Systems Biology, Icahn School of Medicine at Mount-Sinai, and The University of Texas Southwestern Medical Center.
Learn More
Translational Cancer Immunology
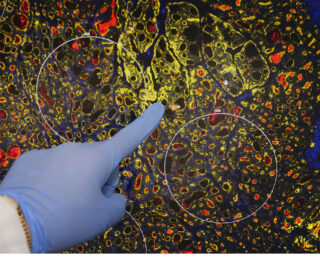
Lead by Dr. Maria Libera Ascierto, the Translational Cancer Immunology department is supported by the Rosalie and Harold Rae Brown Cancer Immunotherapy Research Program. The main objective of the department is to foster Precision Immunology and Biomarker Discovery to investigate how a patient’s immune profile impacts the response and/or resistance to immunotherapy and other therapeutic interventions such has surgery, chemotherapy, and radiotherapy. By doing so, the program aims to better select patients most likely to benefit from immunotherapy and other cancer treatments, thereby helping clinicians detect the insurgence of cancer earlier and select the most appropriate treatment interventions for their patients.
Learn More
Melanoma and Cutaneous Oncology
Lead by Dr. Richard Essner, FACS, Director of Surgical Oncology, the department of Melanoma and Cutaneous Oncology aims to understand the mechanisms of disease metastases and recurrence of skin cancers at the molecular level. With a focus on melanoma, the cutaneous team identifies blood and tumor biomarkers to assess future risk, where findings are utilized to improve outcomes for patients. By developing the tools and indicators that identify development of metastases, we can effectively enhance clinical practices for post-surgical melanoma patients and better support their follow-up care. This promotes enhanced cancer surveillance methods, thereby improving patient survival through early interventions.
Learn More
Sequencing
The Sequencing Center provides a variety of services as a CORE facility, supporting partners such as Illumina (Propel-Certified Service Provider) and HTG Molecular Diagnostics, as part of their Preferred Academic Centers of Excellence (PACE) program. The institute’s services include, mRNA-focused sequencing, targeted resequencing and custom enrichment, OMNI-ATAC-Seq, miRNA seq, ChIP-seq, EPIC 850k Methylation array, and sequencing for pre-made libraries. Our teams have demonstrated proficiency in next generation sequencing at the highest industry standard.
Learn More
Computational Biology
The Computational Biology department is committed to understanding whole systems. The mission is to transform data into biological insights that will ultimately influence clinical practice. The focus is on applying various “omics” data to address translational questions. Current Research Topics include Tumor Epi-Transcriptome, Secondary Cancers, and Metastasis, working to answer specific questions including the role of RNA modifications, molecular differences of cancers based on prior history, and the evolutionary history of tumors from primary to metastasis.
Learn More
Philanthropy Sponsorship
The Saint John’s Cancer Institute has a longstanding history of philanthropic support. Many of these gifts have helped fund groundbreaking cancer research.
Learn More
